EX3: NCI from Molecule and user-defined cube¶
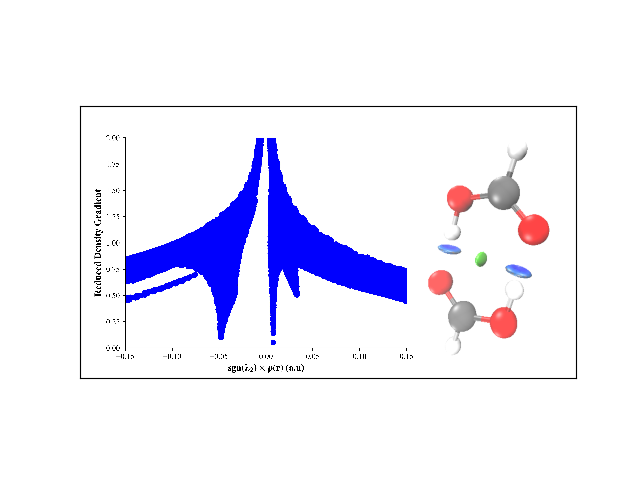
Compute NCI and visualize it for formic acid dimer.
from chemtools import NCI, UniformGrid, Molecule
# 1. Build UniformGrid and NCI model
mol = Molecule.from_file('formic_acid_dimer.fchk')
cub = UniformGrid.from_molecule(mol, spacing=0.1, extension=0.5)
nci = NCI.from_molecule(mol, grid=cub)
# 2. Generate plot, cube file(s) and script for visualizing NCI
# Files generated are formic_acid_dimer-dens.cube, formic_acid_dimer-grad.cube,
# & formic_acid_dimer.vmd
# To visualize the iso-surface, use command: $ vmd -e formic_acid_dimer.vmd
# nci.generate_plot('formic_acid_dimer', denslim=(-0.15, 0.15))
nci.generate_scripts('formic_acid_dimer')
Total running time of the script: ( 0 minutes 11.690 seconds)