EX1: MO from wave-function file and user-defined cube¶
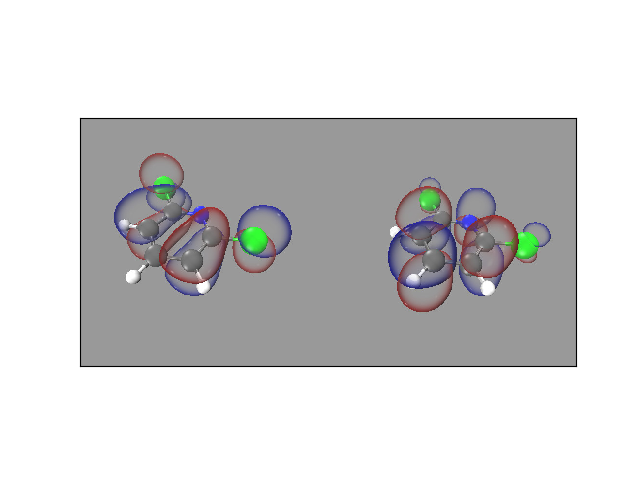
Compute HOMO/LUMO and visualize it for 2,6-dihydropyridine.
from chemtools import MOTBasedTool, UniformGrid
# 1. Build MO Theory model
fname = 'dichloropyridine26_q+0'
mo = MOTBasedTool.from_file(fname + '.fchk')
# 2. Generate cube file(s) and script(s) for visualizing HOMO/LUMO
# Files generated are dichloropyridine26_q+0_mo{index}.cube/.vmd
# To visualize the iso-surface, use command: $ vmd -e dichloropyridine26_q+0_mo{index}.vmd
cub = UniformGrid.from_file(fname + '.fchk', spacing=0.2, extension=5.0)
mo.generate_scripts(fname, spin='a', index=mo.homo_index[0], isosurf=0.025, grid=cub)
mo.generate_scripts(fname, spin='a', index=mo.lumo_index[0], isosurf=0.025, grid=cub)
Total running time of the script: ( 0 minutes 12.228 seconds)