EX7: Dual Descriptor (Frontier MO)¶
Compute dual descriptor on a cubic grid based on the quadratic energy model using frontier molecular orbital (FMO) approach, and generate visualization scripts.
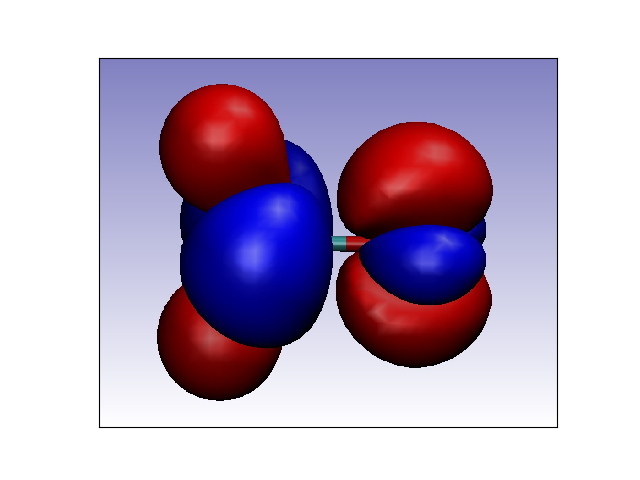
from chemtools import LocalConceptualDFT, UniformGrid, print_vmd_script_isosurface
# 1. Make cubic grid for plotting dual descriptor.
# The cubic grid points are spaced by 0.2 a.u. & extending 5.0 a.u. on each side.
file_path = 'ch2o_q+0.fchk'
cube = UniformGrid.from_file(file_path, spacing=0.2, extension=5.0)
# 2. Build quadratic energy model for Formaldehyde using FMO approach.
tool = LocalConceptualDFT.from_file(file_path, model='quadratic', points=cube.points)
# 3. Dump dual descriptor evaluated on cubic grid.
cube.generate_cube('coh2_dual_fmo.cube', tool.dual_descriptor)
# 4. Generate VMD scripts to plot dual-descriptor iso-surface.
# To visualize the iso-surface, use command: $ vmd -e coh2_dual_fmo.vmd
print_vmd_script_isosurface('coh2_dual_fmo.vmd', 'coh2_dual_fmo.cube', isosurf=0.005,
scalemin=-0.005, scalemax=0.005, colorscheme=[0, 1], negative=True)
Total running time of the script: ( 0 minutes 6.959 seconds)